- Published on
sRNA Analyst
- Authors

- Name
- Ryan Chung
A Comprehensive Tool for In-depth Analysis of sRNA-seq Data across Conditions and Target Genes.
Website: https://cosbi7.ee.ncku.edu.tw/sRNAanalyst
Github: https://github.com/RyanCCJ/sRNAanalyst
Docker: https://github.com/RyanCCJ/sRNAanalyst-docker
What is sRNA Analyst?
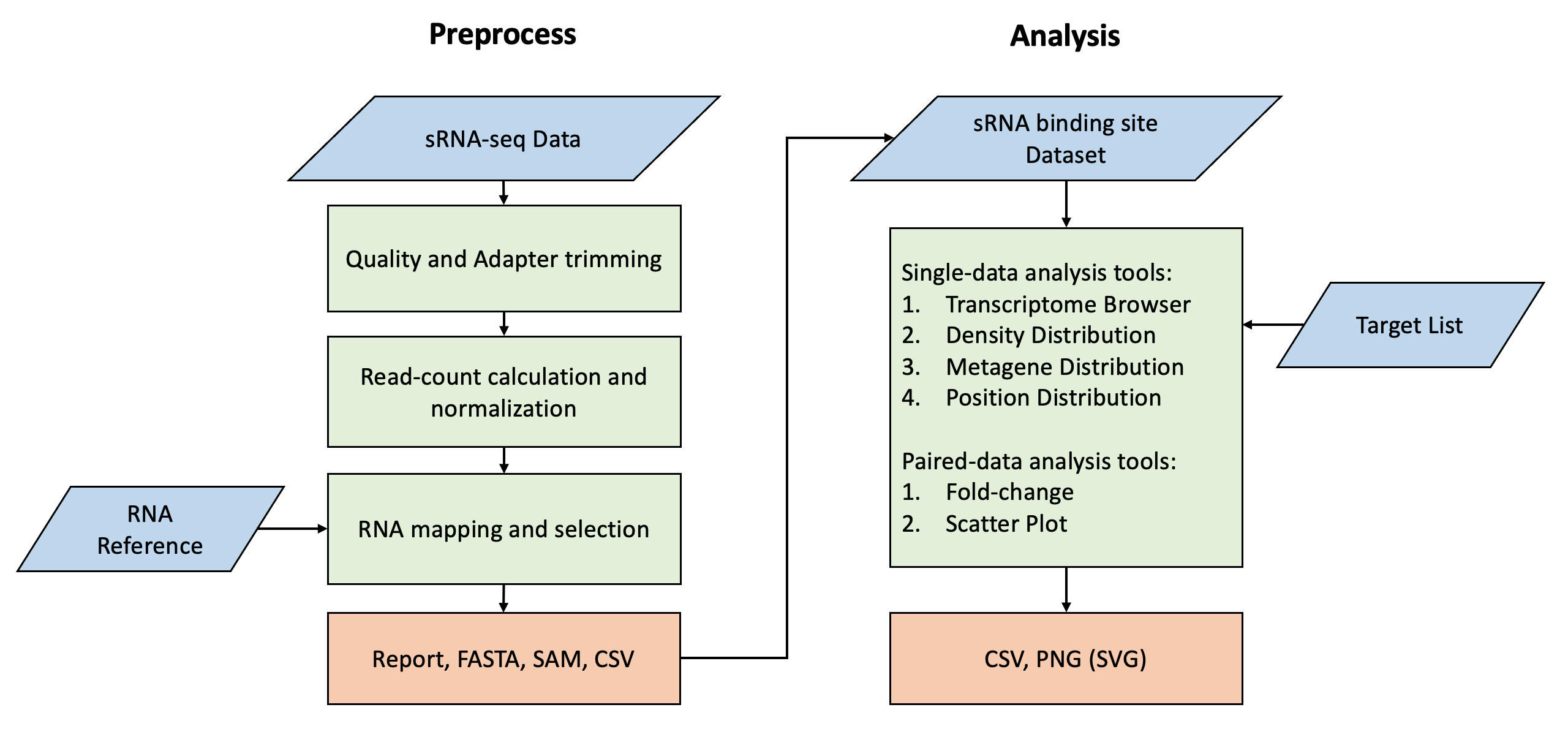
sRNA Analyst is a web-based analysis tool optimized for sRNA-seq data, including a complete data preprocessing workflow, downstream analysis tools, and a collection of sRNA references and experimental datasets.
- The preprocessing tools currently offer a basic workflow for small RNA, including adapter trimming, quality control, length filtering, normalization, and reference alignment. It provides additional helper tools for processing reference files or converting a SAM/BAM file to CSV format for downstream analysis.
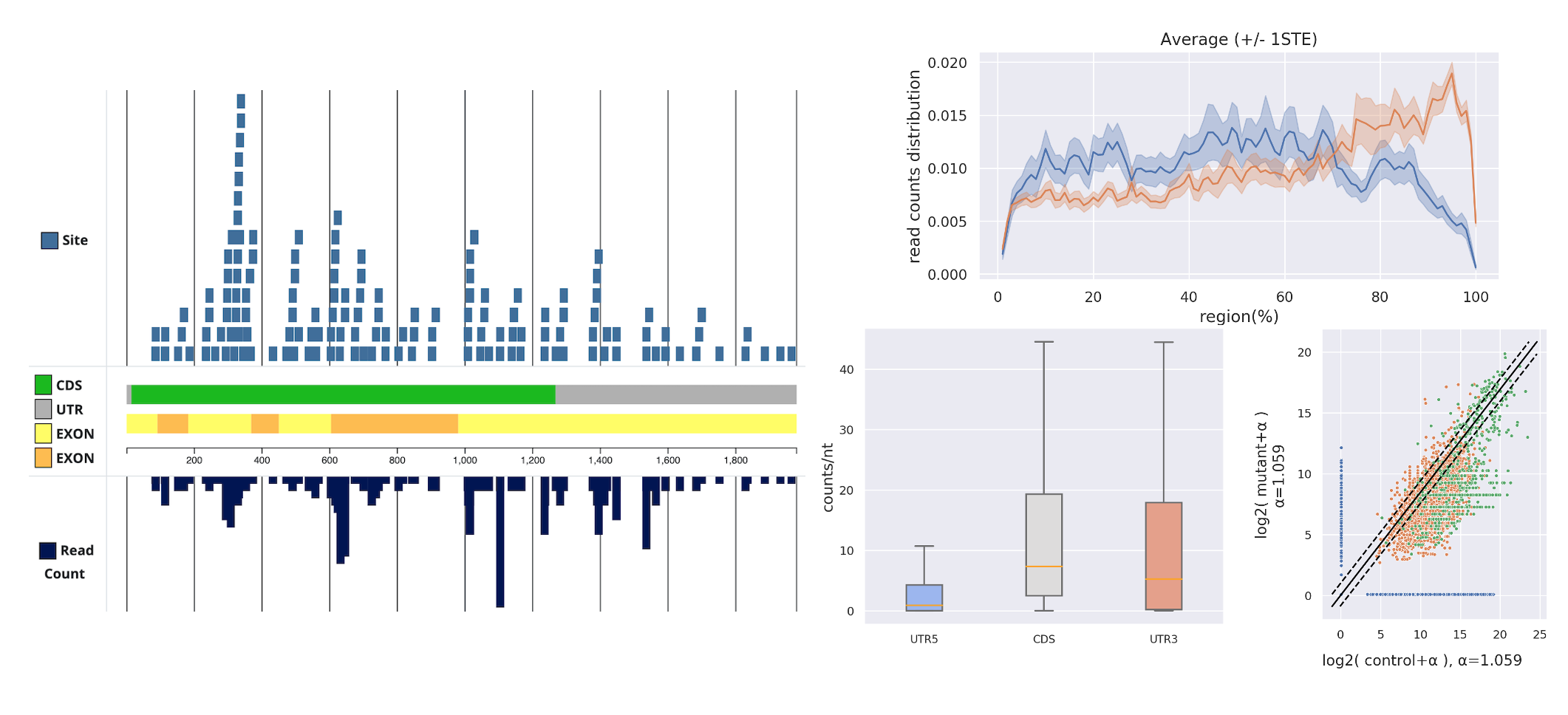
- The downstream analysis tools include Transcriptome Browser, Metagene, Region Density Distribution, Position (head/tail/start codon/stop codon) Read-count Distribution, Fold-change, and Scatter Plot, assisting in observing the distribution of small RNA on specific target RNA under different datasets.
- The database contains experimental datasets primarily focused on C.elegans, along with common model organism miRNA, mRNA references, and partial target gene lists of worm, including Soma target, Germline target, WAGO and CSR-1 target.
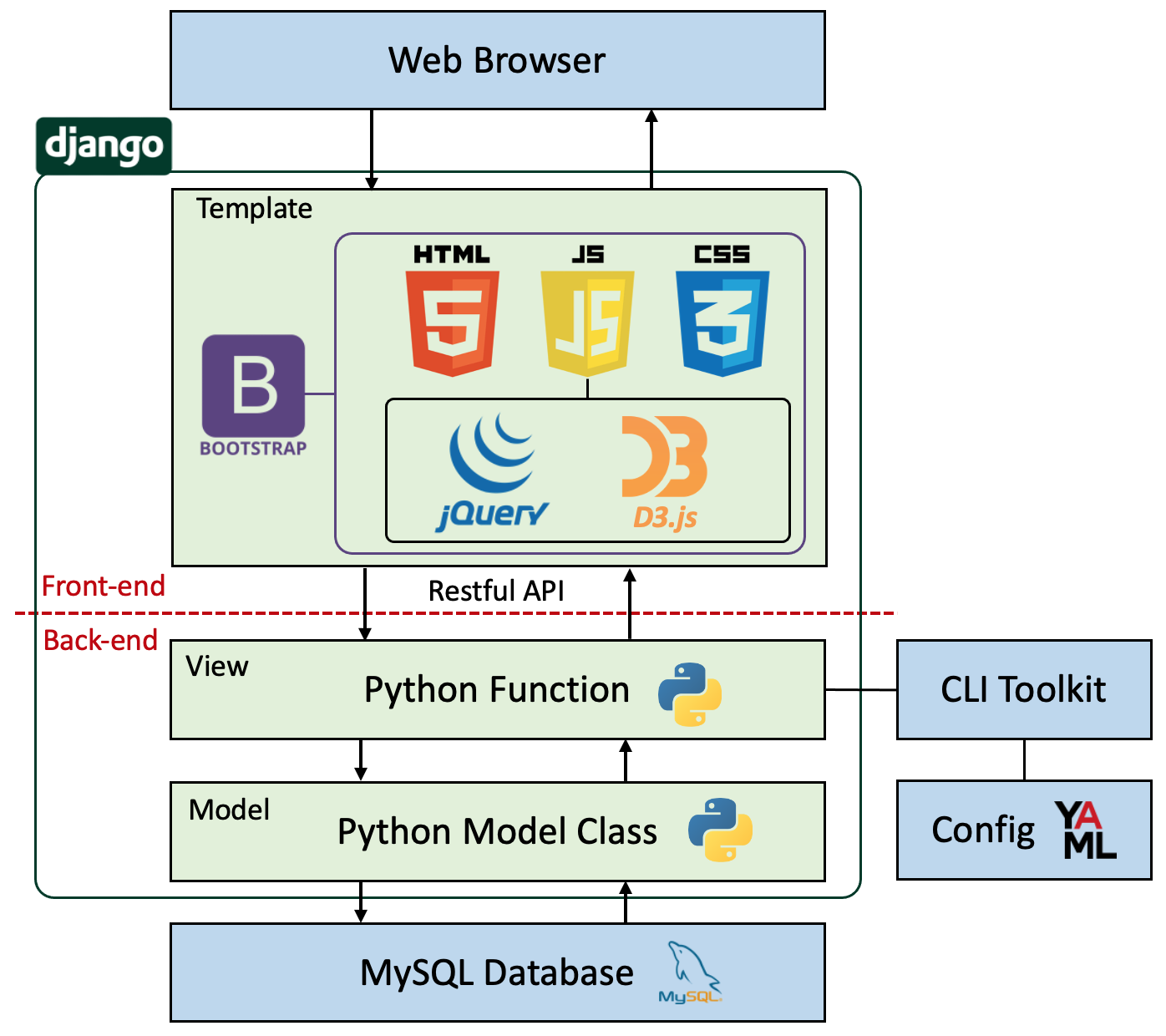
Website Architecture
sRNA Analyst uses the Django MTV (Model-Template-View) architecture, dividing each module and running them independently. The Model serves as the data module, mainly connecting the backend program with the database; the Template acts as the view module, also known as the presentation layer, primarily defining how frontend pages are presented (including HTML, CSS, and JavaScript); the View serves as the control module, mainly handling frontend requests, such as passing form data, performing computational analysis, etc.
The frontend program of sRNA Analyst is primarily written in jQuery and utilizes Bootstrap as the overall template. It uses DataTables to beautify tables and employs D3 to draw dynamic graphics. The backend is developed using Python and R languages, utilizing Shell Script to integrate various preprocessing tools and increasing computational efficiency through parallel processing. The web frontend and backend are connected via Restful API, enabling flexible interaction.
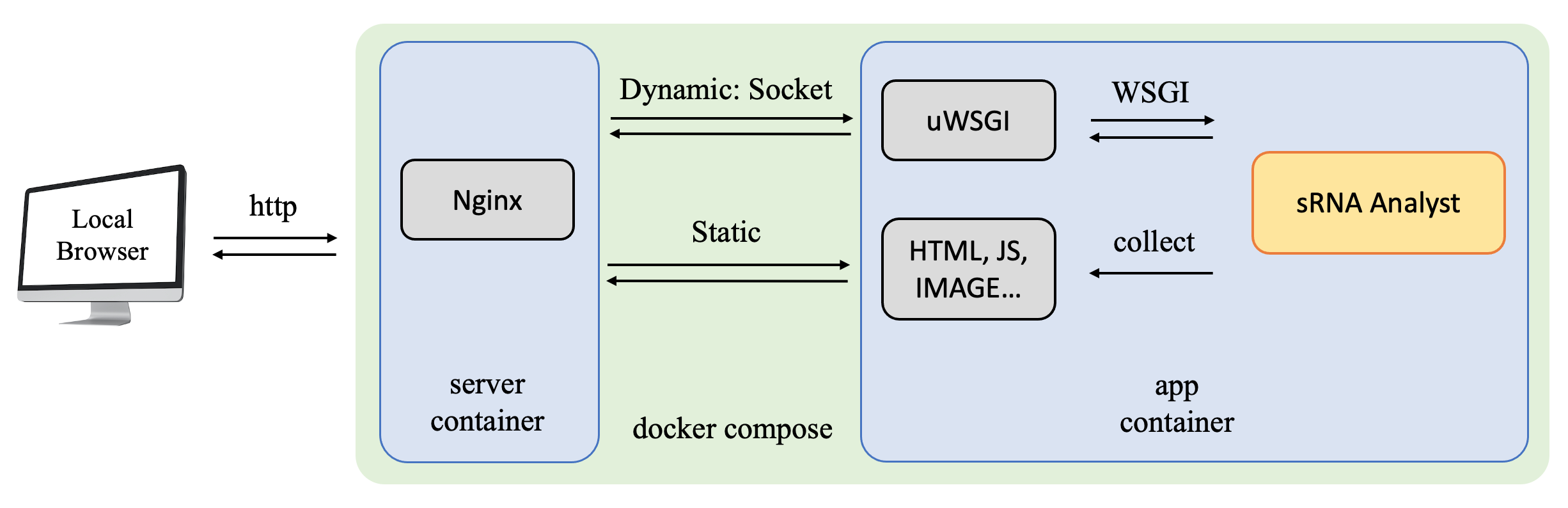
Installation and Usage
sRNA Analyst provides both CLI and Web tools, both of which have Docker versions.
- To use the CLI tool, refer to the documentation on the Github project, install Python and R environments, and you'll be able to analyze NGS data.
- To use the Web tool, refer to the website, and follow the instructions on the Tutorial page to upload data and select analysis tools.
- If network bandwidth and research confidentiality are concerns, you can use our Docker version available on Github and Dockerhub. This allows you to run an Nginx Server locally and use the Web tool in your browser at http://localhost/sRNAanalyst.