- Published on
RNA-seq Analysis Toolkit
- Authors

- Name
- Ryan Chung

CLASH and CIMS
CLASH is an important experimental method for studying RNA-RNA binding. Initially, we induce UV crosslinking between proteins and RNA through ultraviolet irradiation. Subsequently, excess portions are degraded enzymatically. Then, we ligate regulator RNA with target RNA and extract the target protein-RNA complex through immunoprecipitation.
However, during the immunoprecipitation and protein degradation processes, protein residues may be generated at the crosslink sites. This can lead to mutations at specific points when RNA is reverse transcribed into cDNA. These mutations could be deletions, substitutions, or truncations, and their effects will also be reflected in NGS sequencing data, also known as Crosslink Induced Mutation Sites (CIMS).
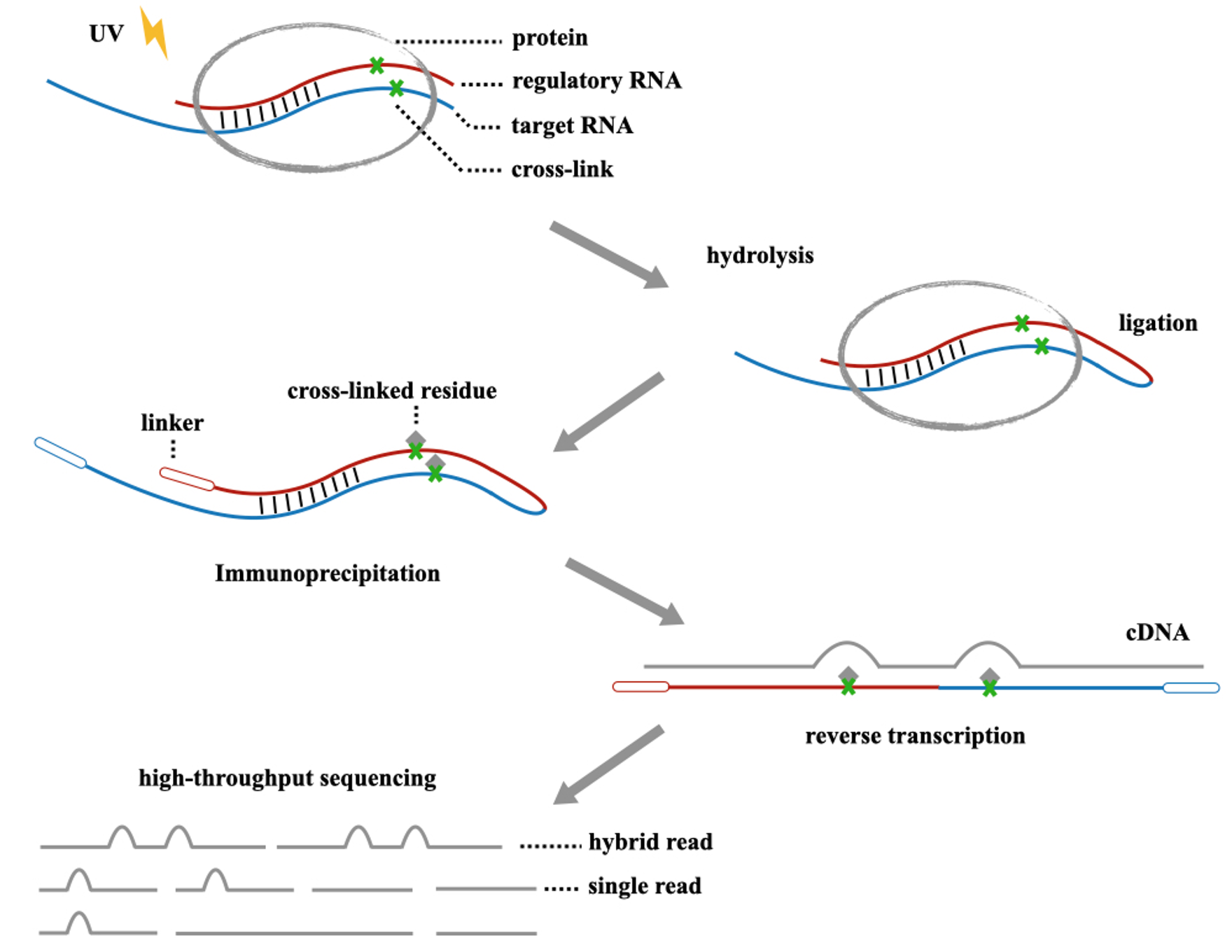
MutaCLASH
Github: https://github.com/RyanCCJ/MutaCLASH
We believe that by studying CIMS, we can infer the true binding sites of proteins and RNA. Therefore, we develope a tool called MutaCLASH. It provides a comprehensive analysis pipeline for identifying mutation sites and binding sites in hybrid reads derived from CLASH experiments.
MutaCLASH features include:
- Uses CLASH Analyst to preprocess NGS data.
- Uses HYB, CLAN and ChiRA to identify suitable hybrid-reads.
- Detects mutation information using Bowtie2 and BWA.
- Utilizes algorithms such as pirScan, miRanda, and RNAup to identify binding sites.
- Generates visualizations of the distribution of mutations.
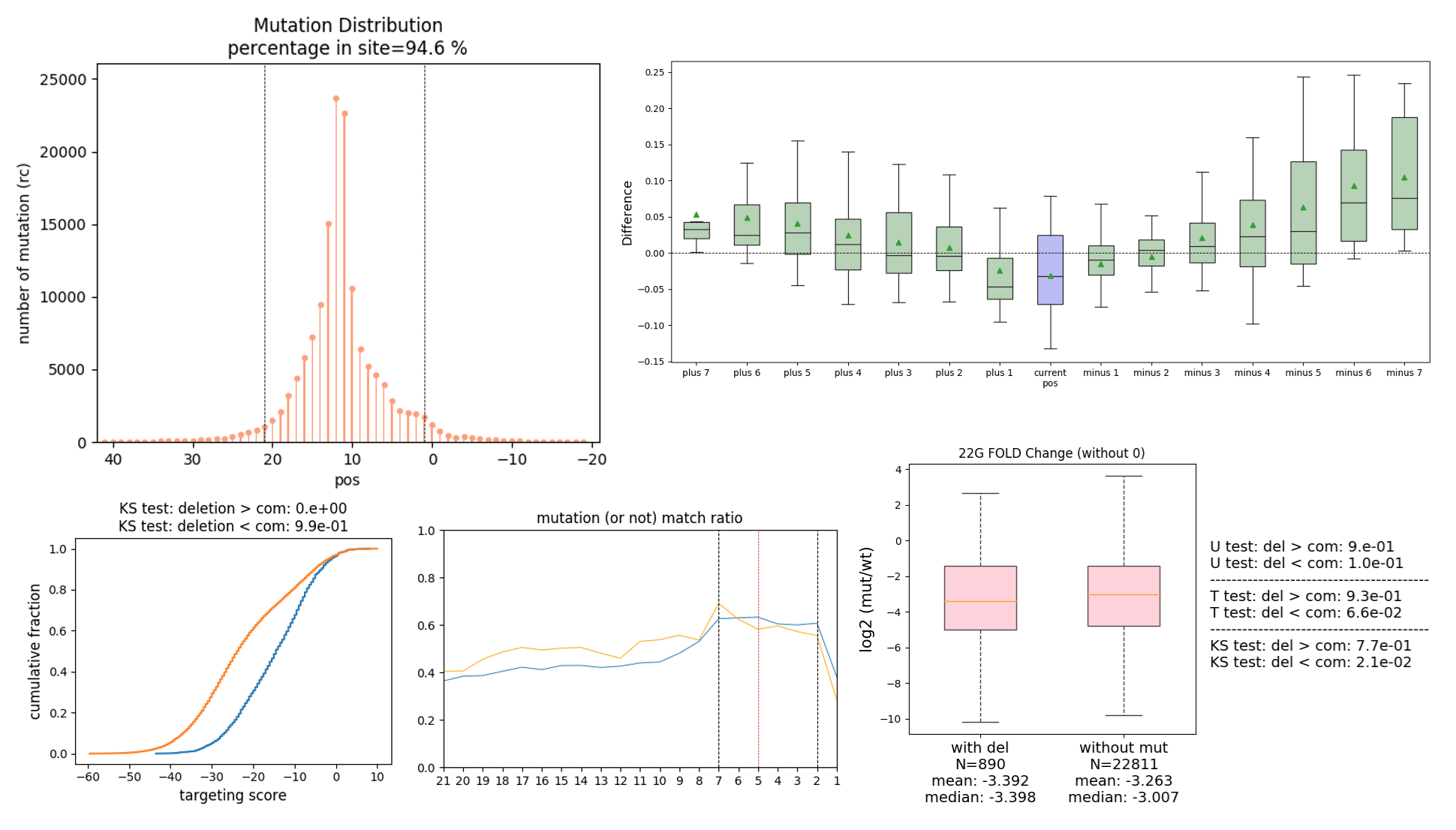
The analysis pipeline can automatically complete all steps within a single command. To see full documentation, please check our Github.
RNA-seq Read Distribution Toolkit
Github: https://github.com/RyanCCJ/RDT
New Project: https://github.com/RyanCCJ/sRNAanalyst
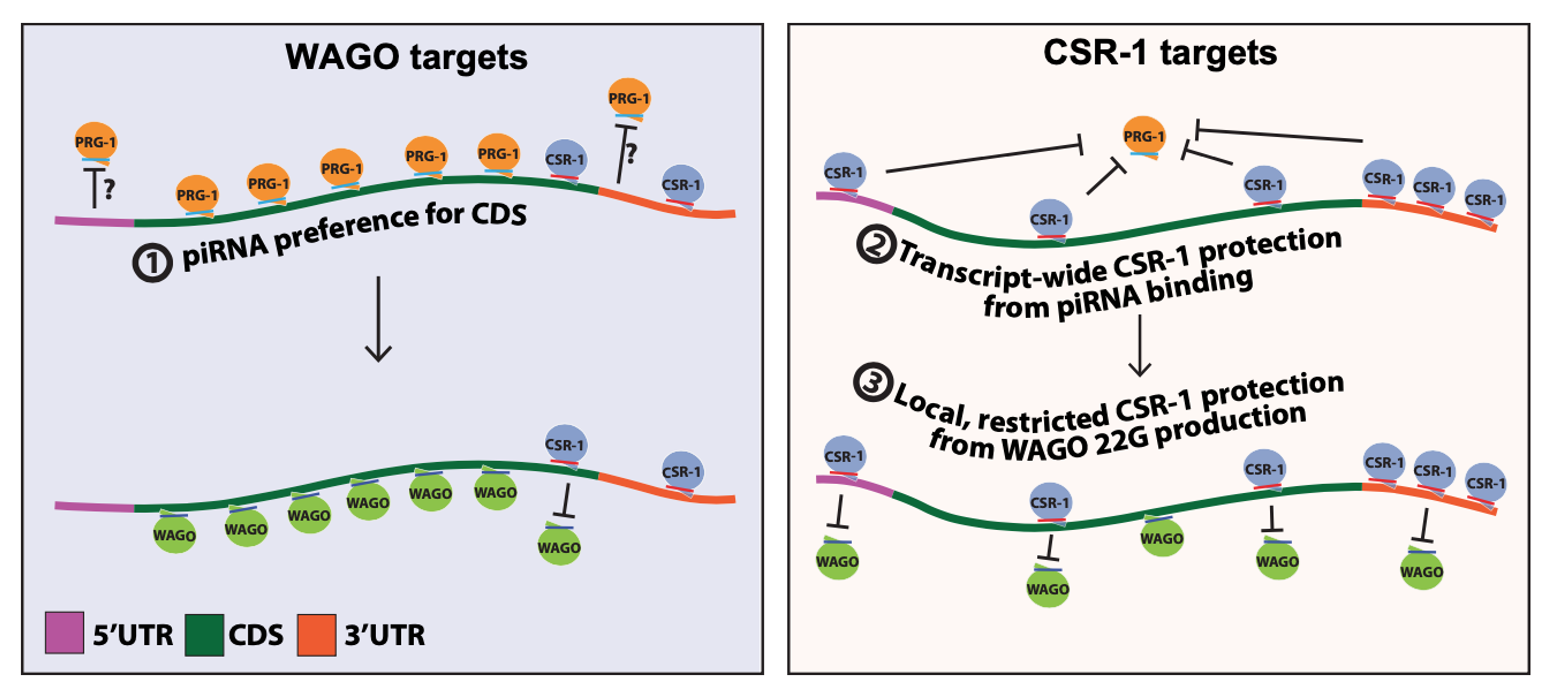
To investigate the binding preferences of regulator RNA on target RNA, we have also developed some user-friendly tools to assist in observing the variations between different target RNAs (such as WAGO and CSR-1 targets) in different regions (such as UTR and CDS).
Additionally, we have integrated data preprocessing functionalities and streamlined the entire NGS research pipeline into a new project, reducing the barrier of entry for biologists in the form of a web-based tool.
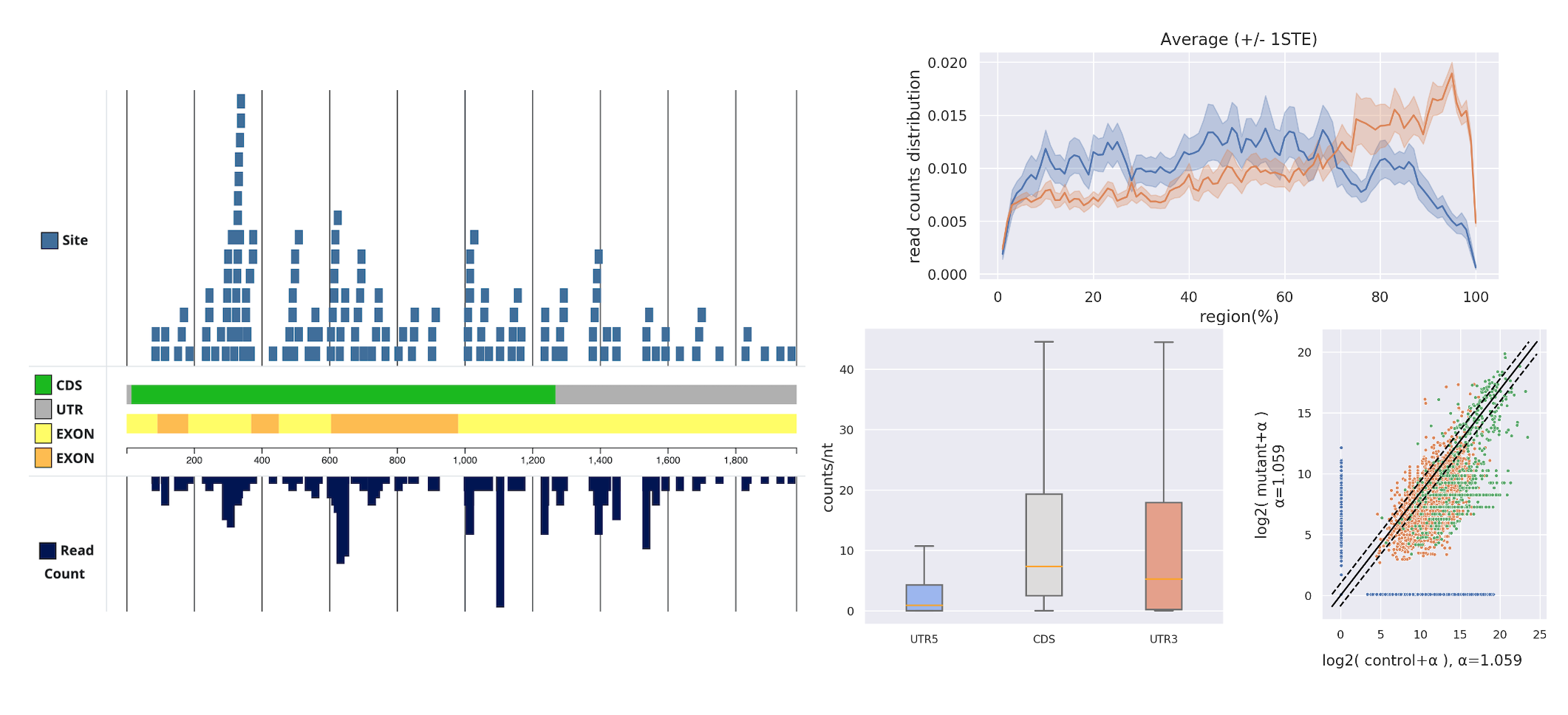
In collaboration between our laboratory at NCKU and UChicago, we analyzed and published a journal paper about the piRNA binding preferences of C. elegans using this tool. 1

Footnotes
Wu WS, Brown JS, Shiue SC, Chung CJ, Lee DE, "Transcriptome-wide analyses of piRNA binding sites suggest distinct mechanisms regulate piRNA binding and silencing in C. elegans," RNA, May 2023, vol. 29, pp.557-569, doi: 10.1261/rna.079441.122. ↩